This article requires a subscription to view the full text. If you have a subscription you may use the login form below to view the article. Access to this article can also be purchased.
- Address for Correspondence: Ryan Cordner
, Brigham Young University, ryan_cordner{at}byu.edu
ABSTRACT
Medical laboratory scientists routinely evaluate blood smears in the clinical laboratory. A high level of skill in white blood cell morphology is required to do the task quickly and accurately. Despite significant training and skill, medical laboratory scientists still demonstrate variability and some inaccuracies in their cell morphology skills. In this study, we proposed a series of 7 questions that can guide morphological identification of white blood cells. We validated the potential of this approach with a random forest classifier algorithm using a dataset of 3600 cells. With the 7 categories as input, the random forest model was able to classify white blood cells with an overall accuracy of 98.7%. We further demonstrated that this approach has application for medical laboratory scientists with color blindness by testing a color independent model of white blood cell identification using 5 parameters. Our approach has the potential to improve educational approaches to white blood cell morphology and could increase the consistency of manual differential results between medical laboratory scientists.
INTRODUCTION
The evaluation of peripheral blood smears by medical laboratory scientists has long been a common practice in medical laboratories.1,2 Advances in technology have led to laboratory instruments that can read and classify white blood cells from a peripheral smear before it is reviewed by a medical laboratory scientist.3 Such instruments have reduced the burden of peripheral blood smear review. In the clinical laboratory, it has become common for medical laboratory scientists to only review blood smears that are flagged by an analyzer for abnormal morphology, a practice that was predicted to occur as far back as 1984.4 This practice has resulted in medical laboratory scientists spending the majority of their time reviewing morphologically difficult blood smears. The training and expertise of the medical laboratory scientist have become critical to the accurate identification of white blood cells and the reporting of morphological information from the blood smear.
It has been noted that despite rigorous training and years of experience, medical laboratory scientists can still make mistakes when it comes to reviewing blood smears.5⇓-7 One study evaluated the reporting of variant lymphocytes by medical laboratory scientists. It was found that 31% of medical laboratory scientists reported a different answer when they were shown a cell that they had previously identified, suggesting a high level of intra-individual variability.7 Other work has found that there are various approaches used by morphologists to simplify complex blood smears and that these approaches can lead to inaccuracies. Among the approaches used were framing bias (an approach that looks for features supportive of an already decided conclusion) and availability bias (the act of giving less weight or consideration to answers that are less common).6 The heuristics utilized in this case varied by the individual and led to inconsistencies and inaccuracies that were based largely on the experience and comfort level of the individual in evaluating blood smears. Ideally, standardized approaches to white blood cell morphology could be applied to reduce mistakes and interindividual variability.
Color blindness is another concern that has long complicated the ability of medical laboratory scientists to accurately review a blood smear and to accurately identify white blood cells. Color is a major factor that has been utilized in cell identification, and the concerns with color blindness have been known for some time.8 The prevalence of color blindness in medical laboratory staff has been reported to be between 2.4% and 10%,8,9 and recommendations have been made that medical laboratory staff with color blindness should have their job duties restricted.9 In this study, we have proposed a novel approach to white blood cell identification. We identified a set of 7 questions pertaining to cellular morphology that can guide the approach to white blood cell identification. This model could be used to standardize white blood cell identification between medical laboratory scientists. We validated the potential of this approach using a random forest classifier algorithm. We further showed that this model can be adapted for use by those with color blindness by creating a color-independent model of white blood cell identification.
METHODS
Image Acquisition and Classification
Peripheral blood smear slides taken from the Brigham Young University Medical Laboratory Science Hematology Slide Bank were photographed at 1000× magnification with oil immersion on a Nikon E200 microscope using a Canon Eos Rebel T7i camera. The images were not altered through any digital software after acquisition and were uploaded to an image database for classification. In total, 3600 blood cell images were classified using a 7-parameter model. This model asks an individual to identify the following characteristics in a nucleated cell: granule type, nuclear shape, cellular shape, cytoplasm color, chromatin pattern, granule quantity, and nucleoli quantity. Each cell was classified by a researcher using this method and then verified by a second researcher. The following types of cells were included in the database: segmented neutrophils, band neutrophils, metamyelocytes, myelocytes, promyelocytes, monocytes, promonocytes, lymphocytes, reactive/atypical lymphocytes, prolymphocytes, eosinophils, basophils, nucleated red blood cells, smudge cells, and blasts.
Distributed Random Forest Classifier Algorithm
The distributed random forest algorithm is a classification algorithm that randomly generates decision trees from a given dataset. The decision trees are created through the process of randomly sampling and aggregating the training data. This process is known as bootstrapping. It allows for increased variability in the decision trees produced.10 For each sample, the algorithm will run the data through the preset number of decision trees created in the data training process. The most common response given by the decision trees is used by the algorithm as the correct identification. We coded our distributed random forest using Python 3.9. Our code connected to the H2O Java-based server that would run the distributed random forest algorithm (https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/drf.html). The 3600 previously classified cells were used to train and test the distributed random forest classifier algorithm. Half of the cells were used for training, and half were used for testing of the algorithm. The model was set to create 100 bootstrapped decision trees from the training data. The accuracy of the model was calculated by comparing the results given by the algorithm with the cells that were previously identified by the researchers. Specific model performance metrics were calculated from the testing data and were evaluated for accuracy, misclassification rate, precision (positive predictive value), sensitivity, and specificity. These metrics were calculated as follows:




Here, TP is true positive, TN is true negative, FN is false negative, and FP is false positive. Overall accuracy of the model was determined by averaging the accuracy for each cell identification.
Variable importance was reported by the model based on which parameters led to the greatest reduction in the mean squared error (MSE) of the model. The parameter that led to the greatest MSE reduction was scaled to 1% or 100%, with each variable thereafter graphed in its relation to the highest performing parameter. Area under the curve (AUC) and MSE were reported as calculated by the model.
RESULTS
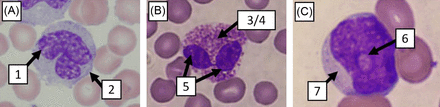
We selected a set of 7 parameters to evaluate white blood cell morphology. The 7 parameters were type of granules (eosinophilic, neutrophilic, azurophilic, etc.), number of granules (none, few, or many), nuclear shape, cell shape, cytoplasm color, presence of nucleoli, and chromatin pattern (Figure 1). Each feature was given a set of predetermined options to describe that feature. Using these parameters, 3600 cells were classified and organized into a database.
Seven-cell morphology parameters. (A) Box 1 indicates chromatin pattern (lacey). Box 2 indicates cellular shape (round). (B) Box 3 indicates granulation type (eosinophilic). Box 4 indicates granulation amount (many). Box 5 indicates nuclear shape (bilobed). (C) Box 6 indicates nucleoli presence (one). Box 7 indicates cytoplasm color (violet).
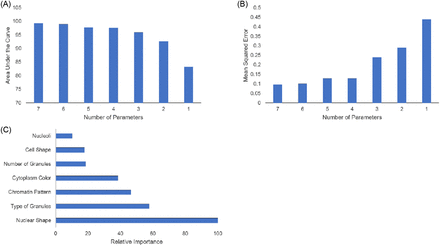
In order to determine whether these 7 features were adequate to accurately identify white blood cells, we trained a random forest classifier algorithm utilizing each of the 7 categories. The model performed very well and was able to identify white blood cells from the categorized data with an accuracy of 98.7%. The overall performance of the model for each cell type was compiled in Table 1. The model performed best in cells that have low variability in their features, such as eosinophils and basophils. The model performed the worst with cells that have high amounts of variability in their features, such as blasts and lymphocytes. The most important features in identifying white blood cells were found to be nuclear shape, type of granules, and chromatin pattern (Figure 2C). These features were found to have the most variance between cell types and thus contributed the most information toward identifying the cells. The least significant identifier was nucleoli presence, which is usually only present in immature cells and reactive lymphocytes.
Cell prediction metrics
Variable optimization and importance. (A) Decreasing the number variables, starting with the least important variable decreased the overall AUC of the model. (B) Decreasing the number of variables used by the model also increased the overall MSE of the model. (C) The relative importance of each variable in reducing the MSE of the model’s performance is displayed. The most important variable is scaled to 100% important.
We next considered whether we could reduce the number of parameters being used and still obtain accurate results. Figure 2A and 2B effectively demonstrated the need for at least 4 parameters to accurately identify cells with an acceptable level of confidence. For our purposes, we considered an MSE below 0.2 as acceptable. Figure 2A illustrates that the AUC decreased as the number of parameters utilized in the model decreased. This also showed that with only 4 parameters (nuclear shape, type of granules present, chromatin pattern, and cytoplasm color) we achieved identifications with 97% accuracy. Figure 2B illustrates the same effect inversely; decreasing the number of parameters that the model utilizes increases the MSE.
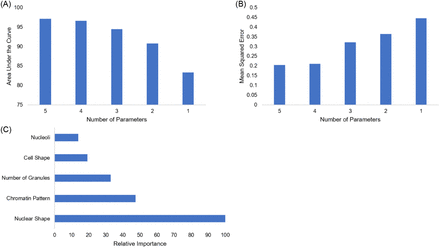
Color has been known to play an important role in identifying white blood cells. We trained and tested the random forest algorithm without color-related data to see if white blood cells could still accurately be identified without this data. Removal of all color-dependent factors used in cell identification (type of granulation and cytoplasm color) yielded an AUC of 97.1% (Figure 3A) and an MSE of 0.2 (Figure 3B). It is important to note that the MSE increased with the removal of parameters in this model faster than it did in the 7-parameter model (Figure 3B). Once again, 4 parameters was probably the least number of parameters that could be used, but 5 parameters yielded better results. The importance of the variables in the color-independent model remained in the same order as the original model (Figure 3C). Our results suggest that the identification of white blood cells can still occur with a high level of accuracy without incorporating color-dependent features.
Variable optimization and importance without color. (A) The impact of decreasing the number of variables, starting with the least important variable on AUC. (B) The impact of decreasing the number of variables used by the model on MSE. (C) The relative importance of each variable in reducing the MSE of the model’s performance is displayed. The most important variable is scaled to 100% important.
DISCUSSION
The results of our study have shown that 7 distinct morphological features can be used to accurately identify white blood cells. The random forest classifier algorithm was able to identify white blood cells with an overall accuracy of 98.7%. We further found that the most important features for identifying white blood cells were nuclear shape, type of granules, and chromatin pattern. Although 7 parameters provided the optimal accuracy, reduced numbers of parameters also yielded actable results. The number of parameters could be reduced down to 4 without leading to an unacceptable increase in the MSE. Below 4 parameters, the MSE was too high to accept the results. A separate study found that using 4 parameters was adequate to have a 76% concordance in identification of various stages of monocyte maturation by morphology experts.11 The 4 selected features were nuclear shape, chromatin pattern, cytoplasm (color and granules), and a miscellaneous comment on general size/appearance. Further work is needed to determine if using fewer than 7 variables is an optimal approach to white blood cell identification.
Using the random forest classifier algorithm, we were able to determine the relative importance of each of the 7 traits for leukocyte identification. These parameters were sufficient to identify cells with a high level of accuracy. The identification of these traits has potentially significant applications in both the clinical laboratory and medical lab science education settings.
In the context of education, the process of learning to identify cells is arduous, often involving trial and error and generally requiring years to achieve mastery. We suggest that our findings could be used to develop a more focused approach to teaching white blood cell morphology. These 7 traits could be taught to students along with their relative importance, and emphasis in teaching could be redirected toward providing students the tools necessary to become proficient in identifying these traits specifically and what about them is indicative of the certain lineages. This focused approach on the relevant cell features could help students follow a common approach and, ideally, could reduce interindividual variability in cell identification. In order to determine the educational merit of this concept, a model for teaching would need to be developed that could be implemented in a classroom. Its impact on time to competency and interindividual variability would need to be evaluated.
In the context of the clinical laboratory, we suggest that an increased emphasis on these traits could minimize disagreement among laboratory scientists. A standardized approach to white blood cell identification would be beneficial for medical laboratory scientists performing morphologically difficult differentials. As those types of differentials have become increasingly the most common type of blood smear reviewed by laboratory scientists, a standardized method in place to identify questionable cells could be very useful. Further work in this context would be required to test a large set of white blood cells from various disorders to ensure the robustness of the approach across various disease states.
Color blindness is a disorder that affects up to 10% of medical laboratory scientists and can negatively impact their ability to perform differentials.8,9 Those who suffer from inherited or acquired color blindness have struggled to accurately identify white blood cells. Educational approaches to help students with color blindness have varied from encouraging them to pick a non–color-dependent career in medicine,12 to recoloring images,13 to teaching histology with grayscale images.14 Although the effectiveness of these approaches has varied, none of them are useful to currently practicing medical laboratory scientists. Our approach to use color-independent features that do not require recoloring or grayscale alterations could be useful in both the educational and clinical settings. In the educational setting, educators would have a framework to use for teaching color-blind students without relying on altered images. Educators would also know which features are important to point out for color-independent cell identification. In the clinical setting, utilizing a color-independent approach to cell identification could reduce the need for screening for color blindness in medical laboratory scientists performing peripheral blood smear differentials and could reduce recommendations that they have their job duties restricted.9
It is important to note that the conclusions of our study are limited to the validation of 7 select morphological parameters as a method that can yield accurate white blood cell identification by a machine learning algorithm. Despite significant potential for the application of this approach, it has not yet been tested in a human cohort. Further research will be needed to determine how our 7-parameter approach could best be implemented in both the educational and clinical settings.
- Received June 10, 2024.
- Accepted September 10, 2024.
American Society for Clinical Laboratory Science