This article requires a subscription to view the full text. If you have a subscription you may use the login form below to view the article. Access to this article can also be purchased.
- Address for Correspondence: Lela Buckingham,
Rush University, lelabarnell{at}gmail.com
ABSTRACT
Colonoscopy provides early detection of colon adenocarcinoma and reduces cancer threat through removal of precancerous adenomas or polyps. Molecular analysis of precancerous polyps could provide information for patient care, eg, scheduling future colonoscopy tests. This study describes potential effects of demographic factors in assessing proposed epigenetic biomarkers (long-interspersed nucleotide element-1 (LINE-1) hypomethylation and genetic mutations) in colon polyps from patients who underwent 2 colonoscopies within 7 years. Polyps from patients for whom cancer or polyps were found in the second colonoscopy were compared with those for whom no lesions were found in the second test. LINE-1 methylation was measured by pyrosequencing and gene mutations were detected by next-generation sequencing. Gene mutations were similar for both groups in a small subset of polyps adequate for sequencing. Among cases with metachronous lesions, 36% of the index polyps (polyps from the first colonoscopy) had at least 1 gene mutation, whereas 43% of the secondary polyps had gene mutations. LINE-1 methylation was significantly higher in polyps collected from patients with metachronous lesions (P < .001); however, confounding influences of demographics were observed. The results of this study demonstrate the potential of assessing molecular biomarkers in polyps with consideration of patient characteristics to make prognostic predictions.
INTRODUCTION
An estimated 104 610 new diagnoses of colon cancer will occur in 2020, with 53 200 deaths attributed to colon and rectal cancer. Colon cancer is most often found in people aged 60 years or older and cancer screening has been strongly promoted within the last few years. According to the Center for Disease Control, colon cancer screening in adults aged 50 to 75 years increased by 4.2 million between 2016 and 2018.1 A variety of screening methods for molecular markers including fecal DNA testing and circulating nucleic acid testing are now available.2 Preventative screening for colon cancer is considered an underutilized tool, even though colon cancer is the second most common cause of cancer death in the United States.1 The importance of screening is reflected in the 5-year patient survival rate being directly linked to the tumor stage at time of diagnosis.3
Laboratory tests that detect changes to DNA structure, sequence, or gene expression (epigenetics) from stool samples or blood are emerging as useful methods when screening for intestinal lesions.4 Detection of somatic mutations in proto-oncogenes, such as KRAS, NRAS, or BRAF in tumor tissue, can be assessed through routine laboratory methods including pyrosequencing and next-generation sequencing (NGS).5 Currently colonoscopy is the most frequently used screening tool for early diagnosis and colon cancer prevention. This procedure offers treatment benefit by removal of polyps or adenomas in the course of the screening. Identification of molecular changes in this precancerous tissue may contribute valuable prognostic information before cancerous tumors arise.
Epigenetic factors, such as DNA methylation, also have the potential to aid in precancerous detection and prognosis. DNA hypermethylation of regions surrounding genes, particularly tumor suppressor genes, alters gene expression patterns in cells. When a tumor suppressor gene loses function as in the case of promoter hypermethylation, the protective effect of gene regulation is lost.4 Decreased methylation (hypomethylation) of noncoding sequences between genes can cause structural abnormalities in chromosomes. Oncogene activation through chromosomal instability promotes unregulated cell division, cell proliferation, and growth. Quantitative methylation changes have been observed, not only in malignant tumors, but at early stages of precancerous lesions.5
Loss of the heavy methylation in long-interspersed nucleotide element-1 (LINE-1) promoters has been observed in cancer cells and therefore is another promising biomarker for the assessment malignancy or potential malignancy.6 Roughly half the human genome consists of repeated sequences including mobile interspersed repeats (transposons and retrotransposons).7 Retrotransposons replicate through an RNA intermediate and are universal throughout the genome. The most abundant retrotransposon is LINE-1, making up approximately 17% of the human genome. Heavy methylation of LINE-1 promoters inhibits transcription. Silencing of LINE-1 elements protects chromosomal stability that would be lost upon their movement through transcription of their RNA intermediates. Linear relationships have been established between hypomethylation of LINE-1 and colon cancer aggression and mortality.8,9
Genetic and epigenetic components have been assessed in malignant tissue; however, less has been applied to precancerous lesions. Adenomatous tissue (polyps) detected and removed by colonoscopy may hold genetic and epigenetic prognostic information for future screening decisions such as development of liquid biopsy methods. Polyps may be present at the time of cancerous tumors (synchronous). Polyps and tumors may also occur at different times of assessment (metachronous). It has been suggested that synchronous and metachronous polyps and tumors might represent similar disease entities with different courses.10
Our laboratory has previously observed significant LINE-1 hypomethylation in polyps removed with synchronous cancer compared to polyps removed in the absence of concurrent malignancy.11 The study reported here was performed to investigate LINE-1 promoter hypomethylation and genetic mutations in polyps as biomarkers for future colon lesions, and the influence of demographic factors on these predictions. The polyps tested were from patients with or without tumors on subsequent colonoscopies (metachronous lesions) at least 5 years after the initial (index) polyps were removed.
METHODS
Patients and Samples
Sixty-five specimens of index polyps from the Pathology Department of Rush University Medical Center were used for this study (IRB #12121202). All polyps were tubular adenomas fixed in 10% buffered formalin. About half of the patients (n = 35) did not develop further colon lesions within 7 years of the index colonoscopy. One patient has since had a diagnosis of colon cancer, and 2 patients had gastrointestinal adenocarcinomas (ileum and cecum). All other patients with metachronous lesions had only adenomas in their follow-up testing.
Descriptive statistics are shown in Table 1. Patients who underwent colon cancer–screening colonoscopy were between the ages of 44 and 85 years old, 57% male, 43% female. Less than half (35%) of the patients in this study group were nonsmokers. Assessment of polyp location (left = distal/descending colon, right = proximal/cecum/ascending/transverse colon) showed 37% distal and 58% proximal with a minority of cases having polyps in both locations. Polyp size ≥6 mm was present in 37% of patients.
Demographics vs absence (No) and presence (Yes) of secondary lesions
Histology
Four-micron sections (6–8 per block) were cut from paraffin-embedded polyp tissue blocks and heat-fixed onto glass slides at 55 °C for 60 minutes. One slide from each case was stained with hematoxylin and eosin (H&E). The stained slides underwent review by a pathologist to confirm the adequacy (at least 3 mm2) and type of polyp tissue on each thin section. Diagnostic and demographic data were acquired from pathology reports, histologic evaluation, and chart review. Collected data included gender, age at diagnosis, polyp size, and polyp location (distal or proximal colon).
Macro-Dissection
Using the reviewed H&E-stained slide as a guide, tumor, polyp, or nonmalignant tissue was scraped from 4–5 slides and placed in 60–200 µL lysis buffer (10 mM Tris, 50 mM KCl, pH 8.3, 1.0 mg/mL proteinase K). The number of slides and volume of lysis buffer used were dependent on the amount of polyp available. Samples were incubated for a minimum of 6 hours at 50 °C prior to molecular analysis. Proteinase activity was eliminated at the end of the digestion by a 5-minute incubation at 95 °C.
LINE-1 Hypomethylation
The LINE-1 retrotransposon targeted is located on 22q11-q12, genomic coordinates (GRCh38): 22:15,000,000-37,200,000. The primer sequences were based on repeat elements (locus X58075:111-358). Analysis was based on LINE-1 sequence with GenBank accession number ONS374723. Isolated DNA was converted with sodium bisulfite for assessment of the methylation status of the LINE-1 promoter. Ten microliters of DNA lysate from macro-dissected polyp tissue were bisulfite converted using the Zymo EZ DNA Methylation TM Kit (Zymo Research, Irvine, CA) following manufacturer’s protocol. Sodium bisulfite converts unmethylated cytosines to uracil in DNA. Methylated cytosines are not affected. The converted DNA was amplified using forward primer TTTTGAGTTAGGTGTGGGATATA and biotinylated reverse primer 5’bio-AAAATCAAAAAAATTCCCTTTC. After amplification, 15 µL PCR product was subjected to pyrosequencing on a Pyromark Q24 pyrosequencer (Qiagen Inc) using sequencing primer AGTTAGGTGTGGGATATAGT. The sequence to analyze was TYGATTTTTTAGGTGYGTTYGTTA. The average of the relative percent C (methylated) vs T (unmethylated) at each of 3 CpG sites was reported. Non-CpG cytosines, which should be 100% converted, were included in each sequence to confirm complete conversion.
NGS
The total volume of samples from microdissection of 2–3 tissue sections (depending on the polyp area) were cleaned using spin columns according to the manufacturer’s protocol (QiAmp, Qiagen Inc). Ten nanograms of the cleaned DNA were used for TruSight Tumor15 library preparation as per manufacturer’s protocol (Illumina Inc). Target genes (AKT1, BRAF, EGFR, ERBB2, FOXL2, GNA11, GNAQ, KIT, KRAS, MET, NRAS, PDGFRA, PIK3CA, RET, and TP53) were amplified from the cleaned DNA (5 µL of 2 ng/µL) in two 15-µL reactions per sample using supplied reagents in a nested PCR. The primers used in the first amplification contained binding sites for indexing each sample in a second 35-µL PCR reaction with a second set of supplied primers carrying 8-bp indexes identifying each sample. After indexing, the PCR products (libraries) from the second reaction were bead-purified, quantified by fluorometry (Qubit, Thermo Fisher), and checked by gel electrophoresis. The libraries were pooled, diluted, and denatured. The denatured pooled library was further diluted to 15 pM for sequencing on the MiSeq Sequencer (Illumina) as per manufacturer’s protocol. The reversible dye terminator sequencing run took 27 hours. Base calling and filtering of variants were performed within the MiSeq software (Illumina), producing a final report of variants for each sample.
Statistical Analysis
Basic summary statistics were calculated for percent methylation of the LINE-1 promoter at 3 CpG sites and the average of the 3 sites. Index and secondary lesions were analyzed as dependent samples. The difference in methylation levels among groups was assessed as nonparametric data by Mann-Whitney tests. The median and range of data were displayed in box plots. Paired-sample analysis between the primary and secondary polyps in the patients with secondary polyps on follow-up colonoscopy was assessed through Wilcoxon Sign-Rank. The association of categorical data (dichotomized age and polyp size) was assessed by Chi-Square. These analyses were performed in SPSS statistical software and Microsoft Excel.
RESULTS
Somatic Gene Mutations
To identify somatic mutations, polyp DNA was subjected to NGS. Only samples with sufficient DNA concentration (15 ng/µL) were eligible for NGS testing. Index polyp mutations in BRAF (n = 2), TP53 (n = 5), and KRAS (n = 1) were detected in index polyps with low allele frequency (0.027–0.19).
Five of the 8 index polyps with mutations were from cases with metachronous lesions (Table 2). Among these cases, mutations in primary and secondary polyps were compared. Three cases classed as having no mutations in the index polyps had mutations (two KRAS and one TP53) in polyps from a second colonoscopy. One case with a TP53 mutation in the primary polyp had no mutations in the secondary one, whereas in another case, the primary polyp had a KRAS mutation and the secondary polyp had a TP53 mutation, but no KRAS mutation.
Mutations detected by TST15 tumor panel sequencing
Differences in the mutation states of index and secondary polyps from the same patient would occur if the polyps found in the second colonoscopy arose independently of the polyps found in the first colonoscopy.
LINE-1 Methylation
Based on LINE-1 promoter hypomethylation in tumor cells, LINE-1 methylation level in polyps is a promising biomarker for cellular malignancy. CpG methylation at the 3 sites within the LINE-1 promoter and overall average of the 3 sites are shown in Table 3.
Demographics and percent LINE-1 methylation
LINE-1 methylation levels were not related to smoking status, gender, race, polyp location, nor mutation status in this patient group (Table 3). Categories that showed significant differences in LINE-1 methylation in at least 1 CpG site were metachronous lesions, age, and polyp size.
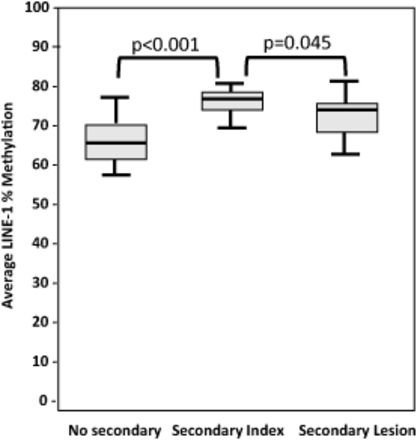
Previous studies have shown significant LINE-1 hypomethylation in polyps with synchronous cancer compared to polyps without.11 In contrast to what was expected from previous studies, LINE-1 CpG sites in polyps with metachronous cancer were found to contain significantly higher methylation than the group without. Median methylation levels (averages of 3 CpG sites in each group) showed statistically higher methylation levels in cases with metachronous cancer (Mann-Whitney U Test P < .001). Figure 1 shows the median and range of the data.
Median of LINE-1 methylation levels of each independent risk group (no secondary average = 64.8%, secondary index = 75.8%, secondary lesion = 68.9%). LINE-1 methylation levels were significantly lower in index polyps without (left) vs index lesion with (center) secondary lesions (P < .001). Paired-sample analysis of secondary index and secondary lesion samples displayed significant LINE-1 hypomethylation in the secondary lesion samples (z = −2.009, P = .045).
To further explore the LINE-1 methylation status in polyps over time, index and secondary lesions were analyzed. Paired-sample analysis of LINE-1 between index and secondary lesions displayed significant hypomethylation in LINE-1 of the secondary lesions compared with the index polyps for each patient (Wilcoxon Sign Rank; z = −2.009, P = .045). Based on previous observations of significant LINE-1 hypomethylation in tumor tissue,8 this result might suggest further progression to a more malignant status in the secondary lesions.
Influence of Demographics
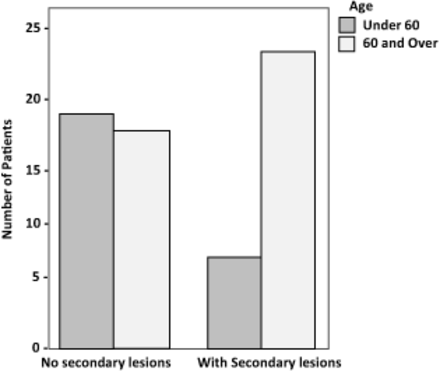
The patient groups were compared with regard to gender, race, age, smoking status, and index polyp location. No bias was identified between the presence of secondary lesions and the descriptive statistics (gender, race, smoking history, polyp location) except for age (Table 3). When patient age was dichotomized into groups (under 60 years, and 60 and over) contingency table analysis (Chi-Square) identified the group with metachronous lesions to contain a significantly older patient population (Chi-Square P = .019; Figure 2).
Total number of participants in each group (with or without secondary lesions) by age (<60 years, and 60 years and older). The group with secondary lesions contained a statistically older patient population (Chi-Square P = .019).
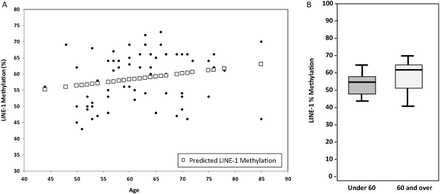
Since the group with secondary lesions contained an older patient population, which could be a confounding factor, LINE-1 methylation was compared to age. Generally genomic methylation increases with age, whereas cancer cells may have lower methylation levels at retrotransposons such as LINE-1. Regression analysis showed a marginal linear relationship between age and LINE-1 CpG site methylation levels (Figure 3A). When average promoter methylation at LINE-1 was compared to dichotomized age, results showed marginally higher LINE-1 methylation in the ≥60 group (P = .105). Figure 3B shows the median methylation levels and ranges of the data.
(A) Regression analysis of age vs LINE-1 methylation. (B) Methylation of LINE-1 with dichotomized age (Mann-Whitney U Test, P = 0.085). Under 60 median methylation = 55%, 60 and over median methylation = 62%.
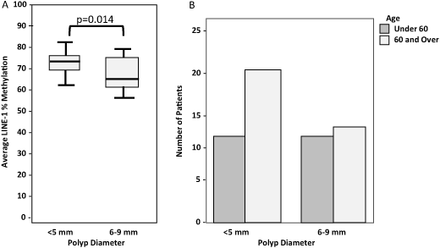
Polyp size may be related to cancer progression. A study of 462 resected sessile and adenomatous polyps showed a stepwise increase in the size of the polyps along with dysplastic progression.12 Another study of patient care based on polyp size found that 7/2115 (0.33 %) small polyps discovered by colonoscopy harbored cancer.13 In the current patient group, polyp size was also related to LINE-1 methylation levels. Larger polyps (6–9 mm) had lower average LINE-1 methylation than smaller polyps (P = .014; Figure 4B). Smaller polyps (<5 mm) were more highly represented in the 60 and over group (Chi-Square P = .019; Figure 4A). This could be a consequence of more frequent colonoscopies in older patient populations.
(A) Smaller polyps were found more frequently in the 60 and over group (Chi-Square P = 0.008). (B) Median LINE-1 promoter methylation compared to polyp size (P = 0.014). Under 5 mm: median methylation = 72%, 6 mm and over: median methylation = 67%.
DISCUSSION
Advances in technology have revealed detailed molecular characteristics of disease states, especially malignant cancers. Molecular changes may appear early in cancer development and some predict disease progression or response to specific therapeutic treatments. These biomarkers then become part of critical laboratory testing.
Patient demographics may influence the use and development of biomarkers based on genetic and epigenetic tumor profiles. Inherited gene variants are associated with younger incidence of cancer, whereas somatic mutations and alterations of DNA methylation develop over time resulting in tumors with somatic mutations and DNA hypermethylation in older patients. Cancer incidence differs with genetics, diet, and other factors.14 Cancer treatment and prognosis are affected by metabolic and hormonal differences.15 Lifestyles and choices such as smoking history or other toxin exposure have well-known associations with cancer. All of these factors could confound identification of biomarkers intended to aid in predicting disease course and treatment strategy.
Genetic variants are well studied in colon cancer and current patient care includes mutational analysis of tumor cells for prognosis and treatment strategy.16 Genetic changes are present in colon polyps; however, most studies focus on inherited mutations and particular polyp types.17,18 Here, BRAF, KRAS, and TP53 variants were found at low allele frequencies in tubular adenomas. The incidence of somatic mutations was 8% BRAF, 4% KRAS, and 21% TP53 in the small number of polyps studied compared to reported highs of 10%–20% BRAF, 50%–70% KRAS, and 55%–70% TP53 in metastatic tumor tissue, depending on primary disease location.19 Five gene mutations were found in polyps from cases with secondary lesions, compared to 3 in cases without; however, due to low sample number, no conclusions as to risk can be drawn. These results suggest that the potential changes in tissue genetic states should be considered in the analysis of genetic biomarkers. Although this small panel does not include all genes associated with colon cancer, the data suggest that TP53 and KRAS genetic changes may occur as early events in the precancerous polyps. No influence of demographics was observed, but this observation is subject to the low sample number.
Paired-sequencing analysis of index and secondary polyps revealed an independent array of somatic mutations in individual patients in the small group tested. The results suggest that polyps found in the subsequent colonoscopy could arise in manners both related to and independent of the index polyps. The limited number of available cases for genetic analysis, however, precludes conclusions with regard to the value of these mutations in polyps as biomarkers. More comprehensive studies would be helpful in revealing the polyp mutation frequency and any association with other demographic factors and polyp characteristics. A recent study using circulating cell-free DNA reported KRAS, BRAF, APC, CTNNB1, FAT3, FAT4, SMAD4, FBXW7, and TP53 genes frequently mutated in colorectal adenoma and concluded that somatic mutations in plasma are potential biomarkers for the diagnosis of colon and rectal cancer.20
LINE-1 promoter hypomethylation is a potentially useful epigenetic biomarker for colon cancer progression or likelihood of recurrence of polyps. DNA hypomethylation in cancer cells has been well established as a source of genetic instability.21 Transposable elements, such as LINE-1, reflect the intergenic methylation state of cells. LINE-1 methylation is decreased in colon tumors compared to normal tissue.22
Despite differences in etiology, paired-sample assessment of LINE-1 methylation levels revealed significant LINE-1 promoter hypomethylation in the secondary polyps compared to the index polyps. Paired-sample analysis (comparing index and secondary lesions within each patient) removes patient-specific demographics as variables. These results, therefore, suggest a potential influence of demographic factors on the predictive value of LINE-1 hypomethylation.
Observation here of increased LINE-1 methylation in polyps from patients with secondary lesions was unexpected, as we had previously observed significantly lower LINE-1 methylation levels in polyps removed from patients with colon cancer compared to polyps from patients who did not have cancerous lesions.11 We have also seen high LINE-1 methylation in normal tissue with progressively lower LINE-1 methylation levels in polyps and lowest in tumor tissues consistent with other studies on such sample sets showing LINE-1 methylation levels decreasing with progression towards malignancy.22 Although it was hypothesized that polyps from patients with risk of secondary lesions in subsequent colonoscopies would show lower LINE-1 methylation levels, the higher levels do not preclude use of LINE-1 as a biomarker. To confirm the use of LINE-1 methylation in polyps or any biomarker, however, several factors must be considered including patient demographics and polyp characteristics.
DNA methylation generally increases with age.23,24 A natural increase in LINE-1 methylation in older patients could affect LINE-1 methylation levels even with the same disease state and prognosis. In the current study, LINE-1 methylation levels were higher in polyps from patients over 60 years of age. The age of patients with lesions found in second colonoscopies was significantly older compared to patients in whom no subsequent lesions were found. Age differences, therefore, could mask hypomethylation as a predictor of disease progression. Associations of lower LINE-1 promoter methylation levels with malignancy may be less strong in older patients.
Characteristics of polyps such as size and location may also denote a progression of dysplasia.25 LINE-1 hypomethylation has been observed in larger polyps (6–9 mm) compared to those <5 mm.26 Although the contribution of polyp size to the prognostic values of molecular biomarkers remains to be determined, the use of markers such as LINE-1 promoter hypomethylation might also take account of polyp size.
To use epigenetic and genetic factors such as LINE-1 hypomethylation and mutational state as biomarkers in colon polyps, a comprehensive and detailed patient history as well as extensive follow-up might be considered. Patient demographics can also influence predictive value of quantitative genetic studies such as cut-point determination for continuous variables. Normal tissue displays a range of LINE-1 methylation levels, which may add difficulty to establishment of a true cut-point between “healthy” and “high-risk” prior to the establishment of malignancy.23
Further studies on adenomas from a large and diverse patient group would confirm observations. Cancerous or precancerous status varied among the patients assessed in this study. Patient acceptance of molecular studies, especially with regard to NGS, might also be considered.27 As patients are under continued evaluation for the presence of colon cancer, there is an opportunity for refined analysis of biomarkers in both paired-sample analysis as well as independent studies.
CONCLUSION
Patient demographics and polyp characteristics can influence the predictive value of genetic and epigenetic biomarkers in precancerous tissues. Patient-specific analysis that obviates demographic differences may better demonstrate relationships between prognosis and biomarker status. Additional studies will further assess how patient demographics and lifestyles influence the use of genetic and epigenetic biomarkers such as mutation status and LINE-1 hypomethylation as predictors for colon cancer development.
FINANCIAL INTEREST
Funding, materials, and equipment were provided by the Department of Medical Laboratory Science, Rush University and the Department of Pathology, Rush University Medical Center.
- Received October 18, 2023.
- Accepted December 11, 2023.
American Society for Clinical Laboratory Science